See Your ChIP-seq Data in a Whole New Dimension
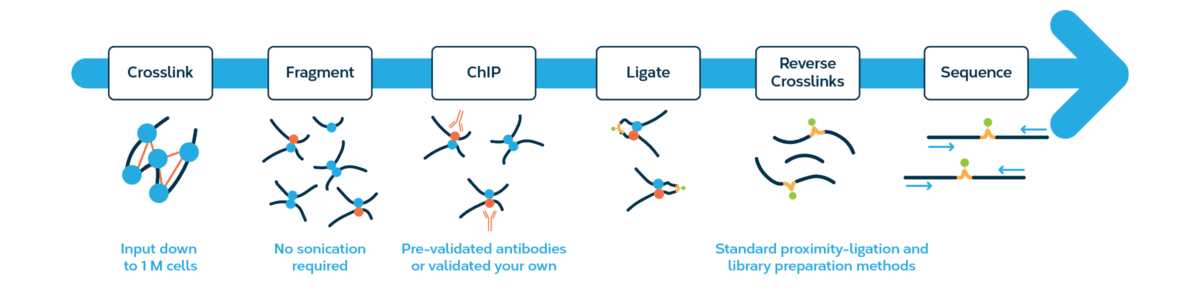
The Dovetail HiChIP MNase Kit combines the benefits of ChIP-seq with Hi-C, a proximity ligation method that captures long-range interactions using standard Illumina paired-end sequencing. Query protein-directed chromatin conformation mediated by specific proteins of interest and see your ChIP-seq data in a whole new dimension.
With a ChIP-seq kit, you only get chromatin information in linear space – meaning you may not be getting all the details you need to gain novel insights on how enhancer/promoter interactions and protein-directed chromatin architecture regulates gene expression.
HiChIP gives your ChIP-seq data a 3D boost so you can take your data to new dimensions. Use conformation contact information to explore how regulatory elements and the spatial relationships between regulatory elements can influence gene expression and drive disease progression.
Nucleosome-Level Resolution
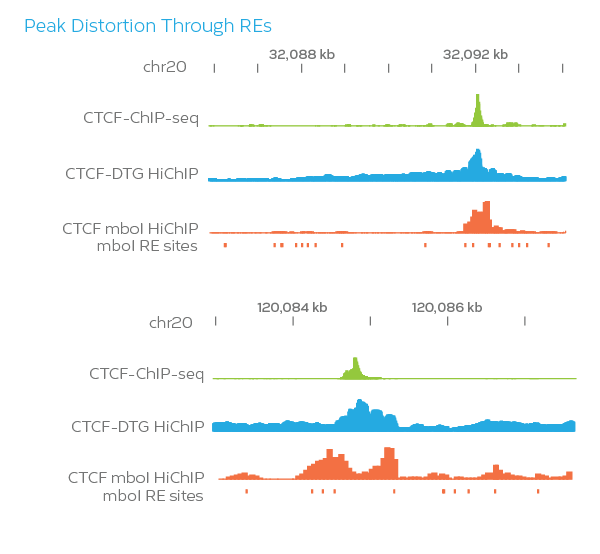
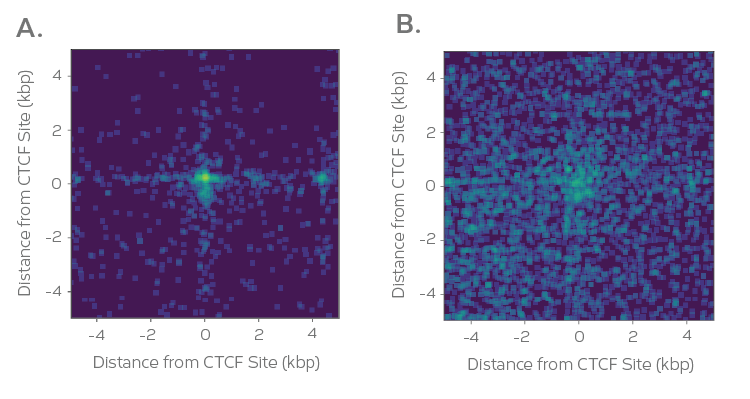
Approximately 20% of the mappable human genome is blind to Hi-C when using restriction enzymes (RE), since signal can be unduly influenced based on the availability of restriction sites close to the protein binding site. The Dovetail HiChIP MNase Kit overcomes this by using micrococcal nuclease (MNase), an enzyme with sequence-independent endonuclease and exonuclease activities, to uniformly fragment chromatin and get the highest resolution 3D genome architecture mapping (150 – 200 bp conformation).
Don’t need ultra-resolution but looking for a way to save of sequencing costs? The Dovetail HiChIP MNase Kit generates data with higher signal-to-noise ratios so you can assess higher chromatin features, such as loop and chromatin interactions, at a fraction of the read depth.
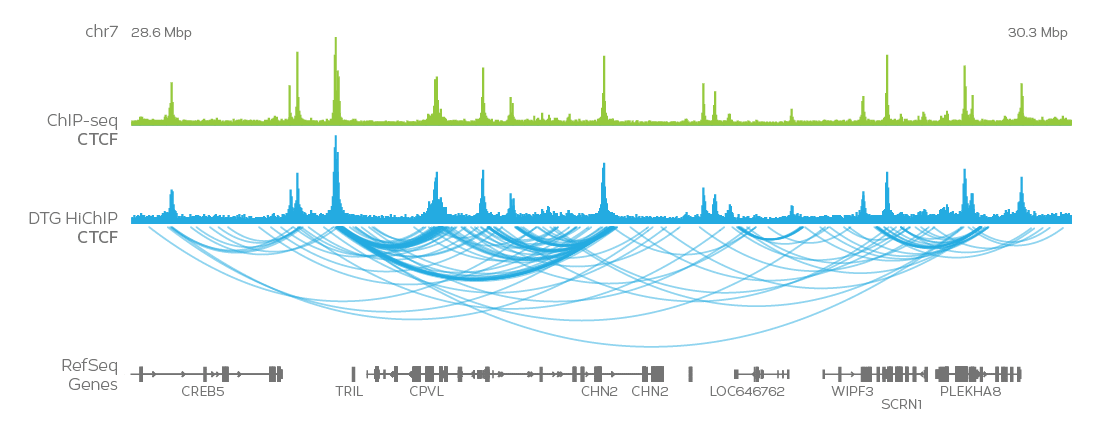
Validated Antibodies
Kit Details
Dr. Orlando Contreras-López – Research Scientist at NGI Stockholm – SciLifeLab
"The quality of service from the Dovetail® scientific support team is one of the best we have experienced from vendors. They show a great deal of technical knowledge and a genuine care in supporting our efforts to prepare high quality proximity libraries using Dovetail® kits."
Webinar Highlight
Our Head of Customer Success, Dr. Myriam Elkhawand, will discuss how Dovetail® HiChIP data enables gene regulation to be understood at a deeper level. During this webinar you'll learn:
The role of genome conformation in gene regulation
Why topological interactions paint a more complete picture of gene regulation
How topology can be used in development, gene regulation, and disease studies
How to combine the 3-Dimensional data of Hi-C with ChIP-seq
-
-
Inferring Protein-DNA Binding Profiles at Interspersed Repeats Using HiChIP and PAtChER
Targeting SWI/SNF ATPases in enhancer-addicted prostate cancer
CTCF mediates the Activity-by-contact derived cis-regulatory hubs
An RNA Polymerase III General Transcription Factor Engages in Cell Type-Specific Chromatin Looping
Loss of Epigenetic Information as a Cause of Mammalian Aging
Case Study: Chromatin Topology Underlies Drug Mechanism of Action
-
-