Dovetail® Omni-C Kit
1000+ Genome Assemblies
Whether you are interested in species conservation, studying the intricacies of evolution or looking to improve the efficiencies of agricultural biology, the first step. Dovetail Genomics continues to be the leading provider of high quality genome assemblies.
With over 900 publications, you can trust our proven kits and services to produce the highest quality, phase informed assembly possible all enabled by Dovetail’s Linked-read technology.
Produce the highest quality genome assembly.
The ultimate scaffolding tool
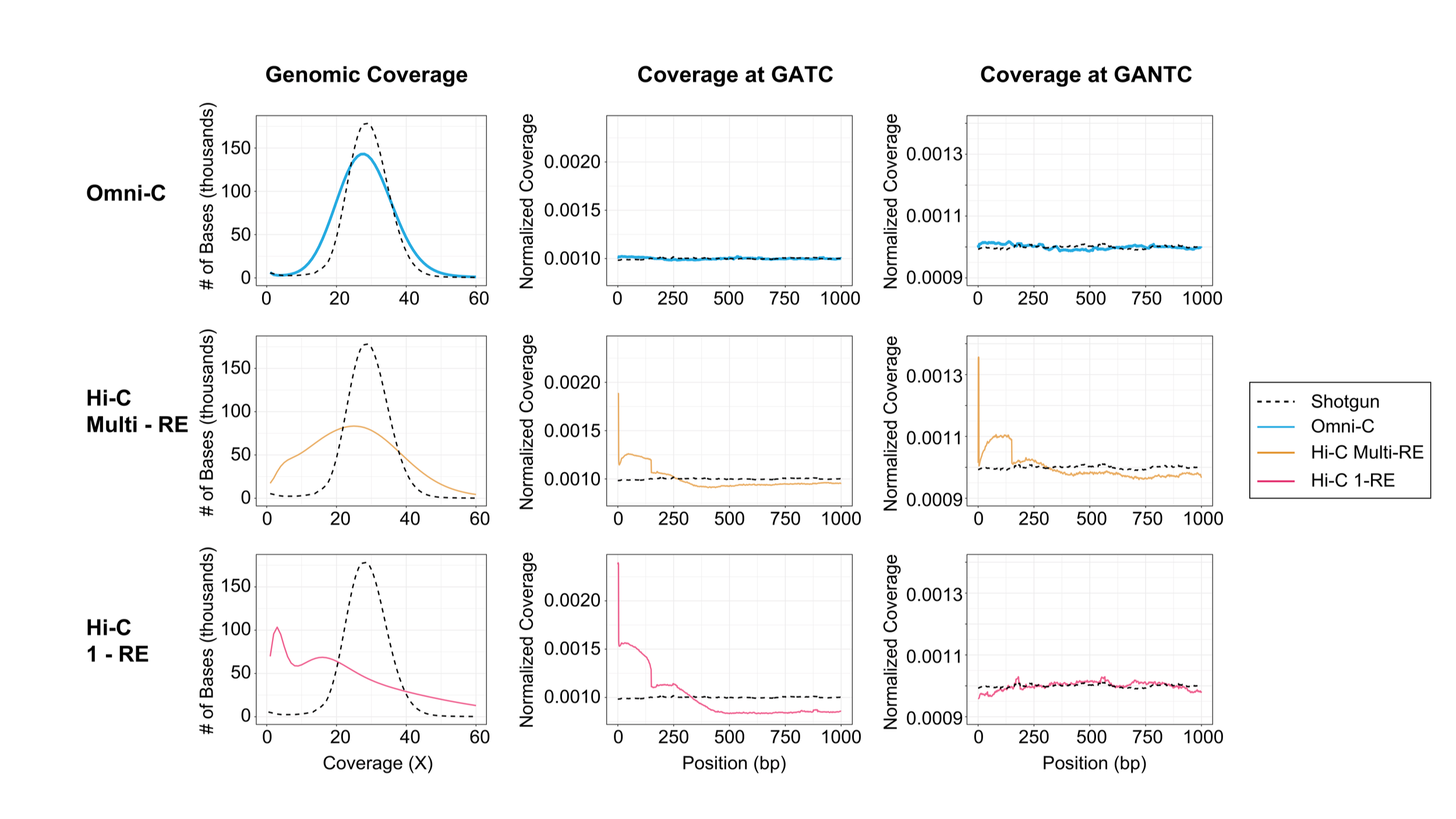
Dovetail® Omni-C® data captures long range interactions while delivering uniform sequence coverage—enabling haplotype phasing
The Omni-C kit moves away from the limitations and bias faced by traditional restriction-enzyme based Hi-C, leveraging the long-range interactions to get a complete picture of the genome.
Omni-C data provides the most complete genome-wide view for all genome assembly, phasing, structural variant, and SNP research questions.
A New Standard: Chromosome scale haplotype aware assemblies made with HiFi and Omni-C technologies
A haplotype-resolved assembly provides a biologically relevant view of your genome. Long-range sequencing approaches, such as PacBio’s HiFi technology, combined with Dovetail's Omni-C libraries make full chromosome scale haplotype-resolved assemblies possible.
Polyploid genomes are among the most difficult to assemble, yet many species of critical importance to evolutionary biology, conservation, and agriculture fall into this category. Because polyploids contain multiple homologous chromosomes, it is often challenging to distinguish between copies. Each gene may be represented by multiple alleles with subtle sequence differences, and resolving these variants with accuracy is essential to linking genotype to phenotype. Achieving this requires additional phasing information during the assembly process.
Dovetail® Genomics overcomes these barriers by combining information-rich Linked-Read data—capturing primary sequence, long-range contacts, and SNP phase information in a single sequencing run—with our state-of-the-art assembly pipeline. This unique approach has enabled us to successfully assemble high-quality genomes for a wide range of polyploid species, delivering the resolution researchers need to advance discovery in complex systems.
Polyploid Assembly
Powering true diploid assemblies
Despite decades of sequencing advancements, many human, plant, and animal reference genomes remain haploid because they lack phase information.
By combining shotgun sequence coverage with the ability to accurately phase SNPs genome-wide, Omni-C powers true diploid (haplotype-resolved) assemblies.
This enriched dataset delivers a more complete view of genetic complexity, enabling investigation of:
Species hybridization
Cis vs. trans mutations
Allele-specific expression
Structural variation
Linkage disequilibrium patterns
Segmental duplications
Complex gene families
Kit Details
-
-
-
See Omni-C Support Page
User Guides
-
Technique: Hi-C
Labeling: Research Use Only
Reactions: 8
Validated Samples
Mammalian cells
Mammalian and non-mammalian animal tissues
Cyropreserved PBMCs
Fresh mammalian whole blood
Nucleated blood
Plants
Application(s)
Genome Assembly, Chromatin conformation analysis, SNP and structural variant detection
Module 1 of 2
Storage
2°C to 8°C
Content
10x Wash Buffer
NWB Solution
Chromatin Capture Beads
Streptavidin Beads
20% SDS
Crosslink Reversal buffer
Module 2 of 2
Storage
-30°C to -10°C
Content
Nuclease Enzyme Mix
10X Nuclease Digest Buffer
100 mM MnCI2
0.5M EDTA
End Polishing Enzyme Mix
End Polishing Buffer
5X Bridge Ligation Buffer
Bridge Ligase
BridgeIntra-Aggregate Ligation Enzyme Mix
Intra-Aggregate Ligation Buffer
Proteinase K
250 mM DTT
HotStart PCR Ready Mix
-
Services
-
The Dovetail® Linked-Read Data powers true diploid assembly through shot-gun sequence coverage and the ability to accurately phase SNPs genome wide enabling the production of fully scaffolded haplotypes. Take advantage of our comprehensive services offerings and the expertise our services team has developed from working with hundreds of diverse taxa.
Reach out to start your project today! -
Polyploids are the most challenging genomes to assemble, yet, many species of great importance to the evolution, conservation and agricultural biology communties fall into this category. Since polyploid genomes contain multiple homologous chromosomes, this makes it challening to distinguish between the different copies. Polyploid organisms can carry multiple alleles for each gene, each differing in sequence. Resolving these allelic variants accurately is critical to understanding phenotype and requires additional phasing information during the assembly process.
Due to these challenges, polyploid genome assemblies have been out of reach for all but the most experienced labs. Even then, the results commonly are unable to capture the full complexity.
Leveraging the information rich Dovetail® Linked-Read data, a datatype that captures primary sequence, long-range contacts and SNP phase information in a single sequencing run, and a state-of-the-art assembly pipeline, Dovetail® Genomics is successfully assembling genomes for a wide assortment of polyploid species.
-
Add annotation for deeper insights
A genome assembly is only as valuable as the biological insights it unlocks. Genome annotation, however, is complex, laborious, and time-consuming—especially without a closely related reference genome. With Dovetail® Genome Annotation Services, you can streamline this step and accelerate your understanding of the genetic blueprint underlying your study.
Explore our genome annotation partnership offering with Goenomics GMBH.