Menu
Menu

Discover the power of Dovetail® LinkPrep™ technology—your new go-to solution for somatic variant detection. With the LinkPrep™ assay, you can easily create linked-read libraries, enabling highly sensitive detection of structural variants (SVs) while capturing the full range of somatic variation. That means one assay delivers SVs, SNVs, InDels, and CNVs with ease.
Best of all, the workflow is streamlined to fit into your day—it takes just a single day from sample to sequence-ready library, requires no specialized equipment, and works seamlessly with any Illumina sequencer. It’s innovation made simple.
The ability to resolve variant phase information is highly limited using standard NGS platforms due to the short reads they produce. While long read sequencing platforms do much better, they are not accessible to all. The lack of variant phase information for many datasets limits their utility in being able to:
• Distinguish compound heterozygotes
• Understand linkage disequilibrium patterns
• Identify disease causing variants
• Determine haplotypes
Dovetail proximity ligation libraries enable short read sequencers to capture long-range phase information alongside traditional genotypes. Break though the limits of your short read sequencer with the Dovetail® LinkPrep™ assay.


Chromosomal large structural rearrangements are associated with a variety of diseases from cancer to rare inherited diseases. These variants can take the form of translocations, inversions, duplications, and deletions. By their very nature, they are challenging to detect using today’s next generation sequencing platforms due to limited read length and continue to be a missing component to most NGS datasets.
The Dovetail® LinkPrep™ assay offers a unique solution enabling the detection of known and unknown structural variants on short read sequencers. The LinkPrep assay for Somatic Variant Detection enables the user to go from sample to sequencer in a single day and is amenable to automation.
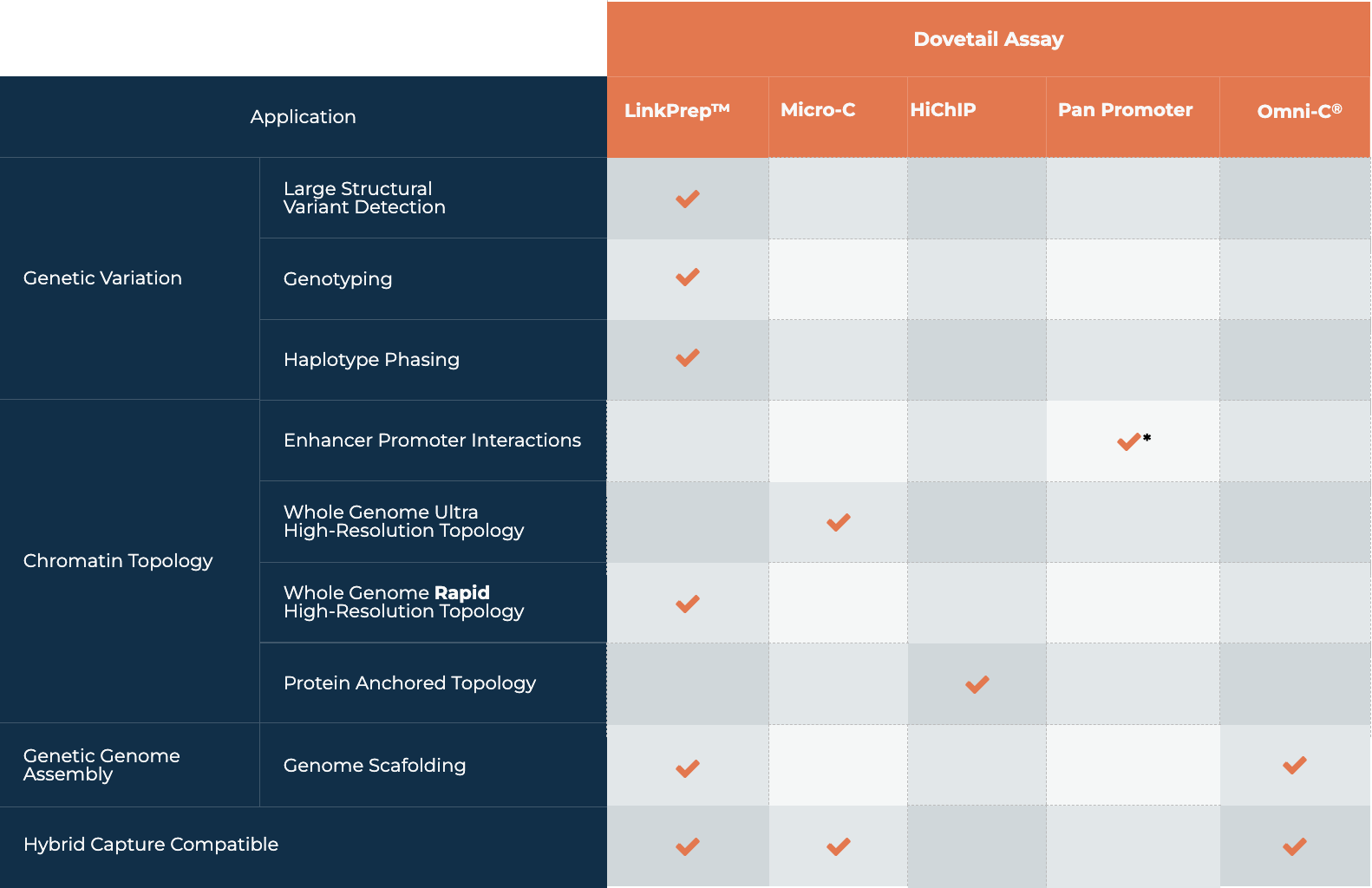
| Application | Use | LinkPrep | Micro-C | HiChIP | Pan Promoter | Omni-C |
|---|---|---|---|---|---|
| Genetic Variation | Large Structural Variant Detection |
 100 Enterprise Way
100 Enterprise Way