Menu
Menu
A recent study led by Phred M. Benham and colleagues has unveiled remarkable findings on the genomes of sparrows, emphasizing the significance of transposable elements (TEs) and the completeness of genome assemblies. This research, published in Genome Biology and Evolution, sheds light on the dynamic nature of avian genomes, challenging previous notions about their stability and repeat content.
The Role of Transposable Elements
Transposable elements, also known as “jumping genes,” are DNA sequences that can move around within a genome. They play a key role in genome evolution by causing structural changes such as insertions, deletions, and inversions, and by influencing gene expression and regulation. While TEs can cause genomic instability, they can also lead to the development of new traits, including color variations and improved immune responses.
Advances in Sequencing Technologies
Previously, the detection and analysis of TEs were limited by short-read sequencing technologies, which struggled to accurately capture repetitive regions of the genome. This created gaps in genome assemblies and hindered our understanding of TE dynamics. However, long-read sequencing technologies, like PacBio and Omni-C, have transformed the field by enabling the creation of highly contiguous reference genomes that better reflect the full scope of TE content.
The Sparrow Genome Study
A recent study focused on producing and analyzing highly contiguous genomes for three species of Passerellidae sparrows: Bell’s sparrow (Artemisiospiza belli), Savannah sparrow (Passerculus sandwichensis), and song sparrow (Melospiza melodia). These new assemblies were compared to other sparrow genomes made using various sequencing technologies. The study found that genomes assembled with long-read technology were significantly more complete and contained more repeat sequences than those assembled with short-read technology.
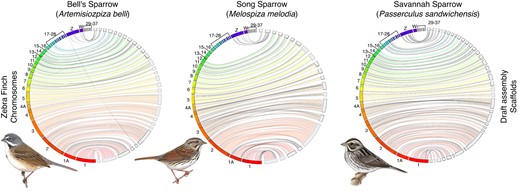
Fig. 1. Jupiter plot comparing higher level synteny and completeness between the zebra finch (Taeniopygia guttata) genome (bTaeGut.4) and each of the three CCGP draft assemblies of Passerellidae sparrow species. Zebra finch chromosomes are on the left in each plot (colored) and sparrow scaffolds are on the right (light gray). Twists represent reversed orientation of scaffolds between assemblies. Song and Bell’s sparrow reference genome samples were both from females, whereas the Savannah sparrow reference was from a male. Song and Bell’s sparrow illustrations reproduced with the permission of https://birdsoftheworld.org with permission from Lynx Edicions. Savannah sparrow illustration contributed by Jillian Nichol Ditner.
Key Findings
Implications for Understanding Avian Genomes
The study emphasizes the importance of using long-read sequencing technologies to capture hard-to-assemble regions of the genome. Creating highly contiguous assemblies allows for more precise TE annotations, offering deeper insights into the evolutionary dynamics of these elements. This is particularly important for bird genomes, which were previously thought to have low repeat content and limited variation.
By enhancing our understanding of TE dynamics in sparrows, this research paves the way for further exploration of genome evolution across various species. The findings also have implications for conservation genomics, providing a more detailed genetic framework for studying population dynamics, adaptation, and speciation in birds. These findings challenge previous assumptions about the stability of bird genomes and highlight the potential of long-read sequencing technologies. As genomic tools and techniques continue to improve, we will gain a clearer understanding of the complex processes driving evolution across the tree of life.
Read the full article here: https://academic.oup.com/gbe/article/16/4/evae067/7639431?login=false
 100 Enterprise Way
100 Enterprise Way