Menu
Menu
De novo genome assemblies have traditionally been pseudo-haploid in nature. Newer, more accurate long read sequencing coupled with unbiased, restriction-enzyme-free proximity ligation technology is enabling high-quality haplotype-phased genome assemblies from a single individual. Phased haplotype blocks are now of chromosome size and are beginning to uncover the true structure of plant and animal diploid genomes.
At Dovetail, we are continually looking for ways to enrich our datasets to provide our customers with genome assemblies that will stand the test of time. Our newest workflow includes PacBio HiFi long reads assembled using HiFiasm1 and Omni-C long range proximity ligation data. The resulting two haplotype-resolved assemblies are then scaffolded to chromosome-scale using Dovetail’s proprietary HiRise software.
Haplotype-resolved assemblies offer many advantages for genomic-based studies in evolution, conservation, agricultural biology, and human disease. In this blog, I will summarize some of the applications where a true diploid assembly offers far more power than a pseudo-haploid assembly.
It is well known that segmental duplications (SDs) play an important role in evolution2. Segments of DNA can be duplicated multiple times in cis or trans and moved around the genome. SDs are typically defined as repeated stretches of DNA greater than 1kb in length and with greater than 90% homology2. They provide genomic redundancy that can be a source of new genetic variation. SDs are recombination and structural variant hotspots and can be a source of very rapid genomic evolution2,3. In humans, SDs account for more variant base pairs than any other type of variation4. For these reasons and more, accurately determining the structure of SDs for both haplotypes is important in many fields of genetic study.

Figure 1. From Megan Y Dennis and Evan E Eichler, 2016. Human adaptation and evolution by segmental duplication, Current Opinion in Genetics & Development, 41:44–52
A prime example of a human haplotype-specific SD event is a 900kb region of chromosome 17 (Figure 1)2,6, a region that has undergone complex inversion and duplication events and includes the MAPT gene, which is associated with neurological disease. The SD event is both population and haplotype specific: Europeans carry both HAP1 and HAP2 while other populations (Asian, African, and Oceanic) only carry the HAP1 haplotype2. There is evidence to suggest that the HAP2 haplotype was introduced into Europe by Neanderthals at least 18,000 years ago5, but interpreting this complex and evolutionary relevant region of the genome requires a haplotype-resolved assembly.
The advantages of acquiring a de novo genome assembly are often underappreciated in the field of conservation biology. The preservation of biodiversity depends on fully understanding the biology of the species under study. A high-quality reference genome is a powerful tool that can be used for better and more efficient species management as it can reveal critical genomic features like admixture, hybridization, runs of homozygosity, amounts of heterozygosity, linkage disequilibrium patterns, and much more. Many of these genomic features are better understood with a haplotype-resolved assembly.
One example of a haplotype-based conservation application is dispersal in European sea bass (Dicentrarchus labrax) populations7. A good understanding of a species’ migration and dispersal patterns is important in conservation and management, and the erosion of admixture tracts over time can be used to infer dispersal distance. Using European sea bass as an example (Figure 2, red arrows), a hybridization zone exists between Atlantic and Mediterranean populations: Atlantic haplotypes are introduced into the Mediterranean population in the Mediterranean Sea, and once there, hybrids disperse away from the contact zone (Figure 2, yellow arrows). Over time, recombination erodes the lengths of admixture tracts. By comparing admixture tract length distributions from Western and Eastern Mediterranean populations, dispersal distance estimates can be quantified7 and used to monitor and manage bass populations.
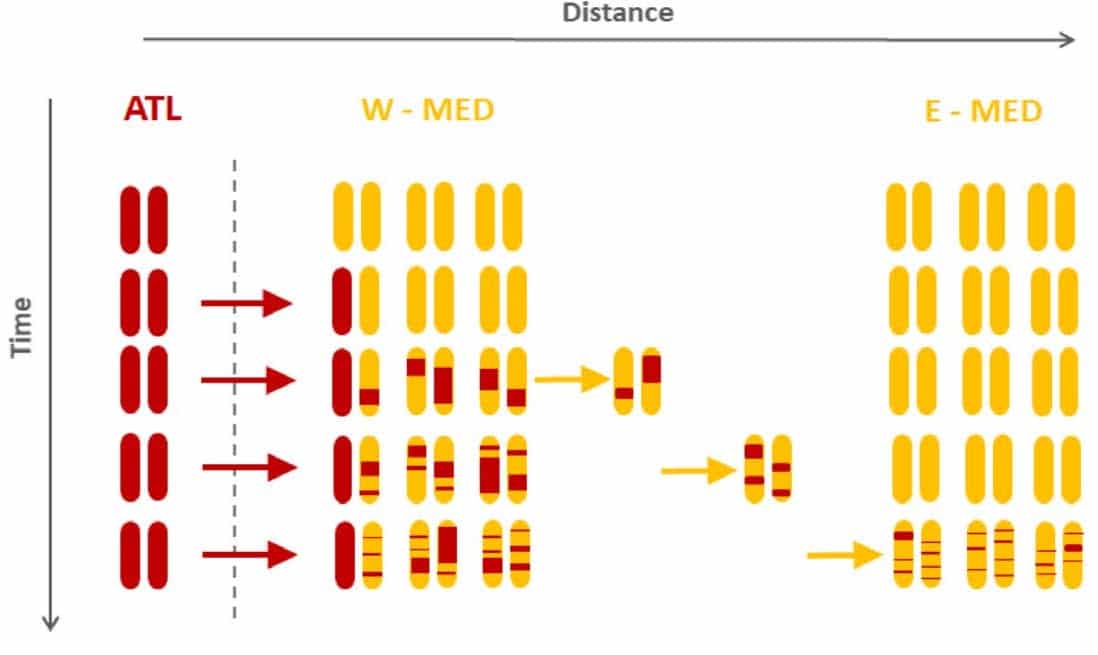
Figure 2. From Duranton et. al., 2019. The spatial scale of dispersal revealed by admixture tracts, Evolutionary Applications. 2019;12:1743–1756
A high-quality haplotype-resolved reference genome is a necessary tool for crop improvement and breeding. Correctly resolving haplotypes on a chromosomal scale will empower the plant biologist with a better understanding of structural variation, hybridization, allele-specific expression, segmental duplications, polyploidy, and much more.
https://dovetailgenomics.com/wp-content/uploads/2022/09/Camellia_sinesis_haplotype-resolved_genome_assembly-1024×450.png
Figure 3. From Zhang et. al., 2021. Haplotype-resolved genome assembly provides insights into evolutionary history of the tea plant Camellia sinensis, Nature Genetics volume 53, pages 1250–1259
Recently, a haplotype-resolved assembly was built for tea (Camellia sinensis)8. Of the 42,628 genes found, 14,691 were biallelic, and some of these genes had significant functional haplotype-specific variations like start or stop codon loss, premature stop codons, or frameshifts; in addition, 87% contained at least one non-synonymous SNP8. Two relevant examples are the tissue and haplotype-specific expression of CsSRC2 and CsGGPS1, which both play an important role in physiology: CsSRC2 is involved in response to cold stress and CsGGPS1 in terpenoid backbone biosynthesis. Moreover, both genes displayed complex haplotype-specific variants that would have not been accurately disentangled with a traditional pseudo-haploid assembly8.
At Dovetail, we are helping to set a new standard in genome assembly quality. Haplotype-resolved assemblies are poised to uncover novel and exciting findings that will empower biologists like never before. Let us know if we can help you build a haplotype-resolved assembly for your species of interest!
“We were blown away by the completeness of the chromosome-scale assembly (Dovetail) produced using HiFi, Omni-C and the HiRise pipeline. With an assembly drastically exceeding Vertebrate Genomes Project standards in hand, we can put all our focus on the downstream science.”
Charles Feigin (Princeton University; received a haplotype-resolved assembly for the Eastern quoll)
Citations
 100 Enterprise Way
100 Enterprise Way