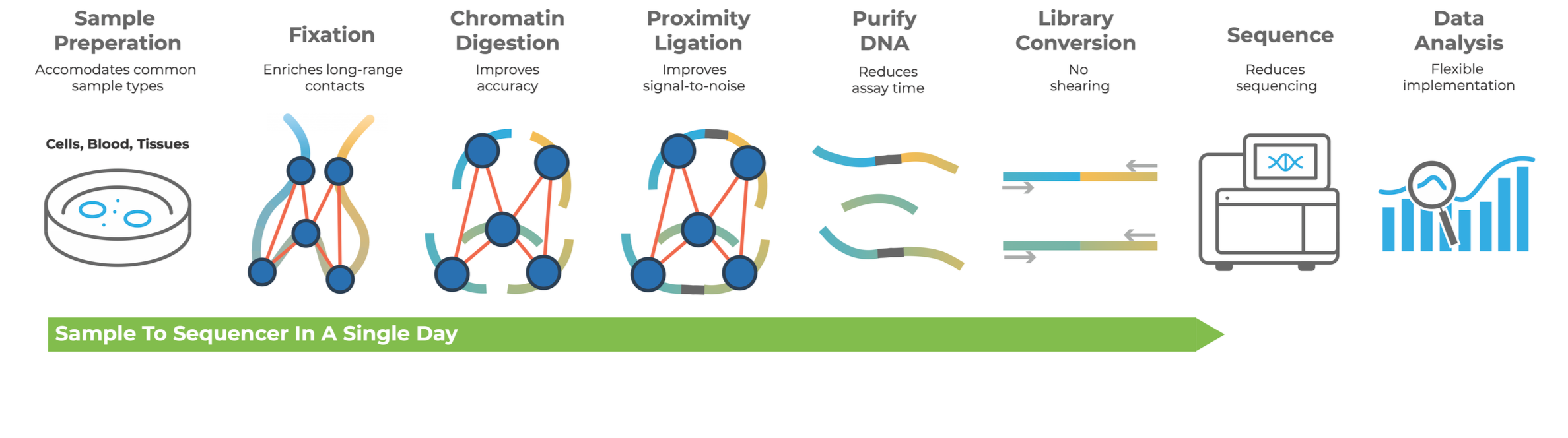
Meet the Dovetail® LinkPrep™ assay
Dovetail® LinkPrep™ technology expands the capabilities of short-read sequencers by capturing long-range genetic information.
With LinkPrep™, researchers can generate high-quality linked-read libraries that capture long-range genomic information, LinkPrep™ unlocking powerful insights across a wide range of applications—including base pair-resolved somatic variant detection, high-resolution 3D genome characterization, and haplotype-aware genome assemblies.
The Dovetail® LinkPrep™ assay features a streamlined workflow that produces sequencing-ready libraries in a single day. Designed for ease of use, the protocol is accessible to users with standard molecular biology training – no specialized expertise required. The LinkPrep™ workflow is automation-compatible and well-suited for high-throughput settings.
Do More with a Single Assay, In Less Time
Broad sample capability, Minimal input requirements
Validated workflows for cells, tissue, and blood. While the standard workflow requires 1 million cells or equivalent, it can also accommodate smaller samples, starting from as few as 10,000 cells, depending on the specific application.
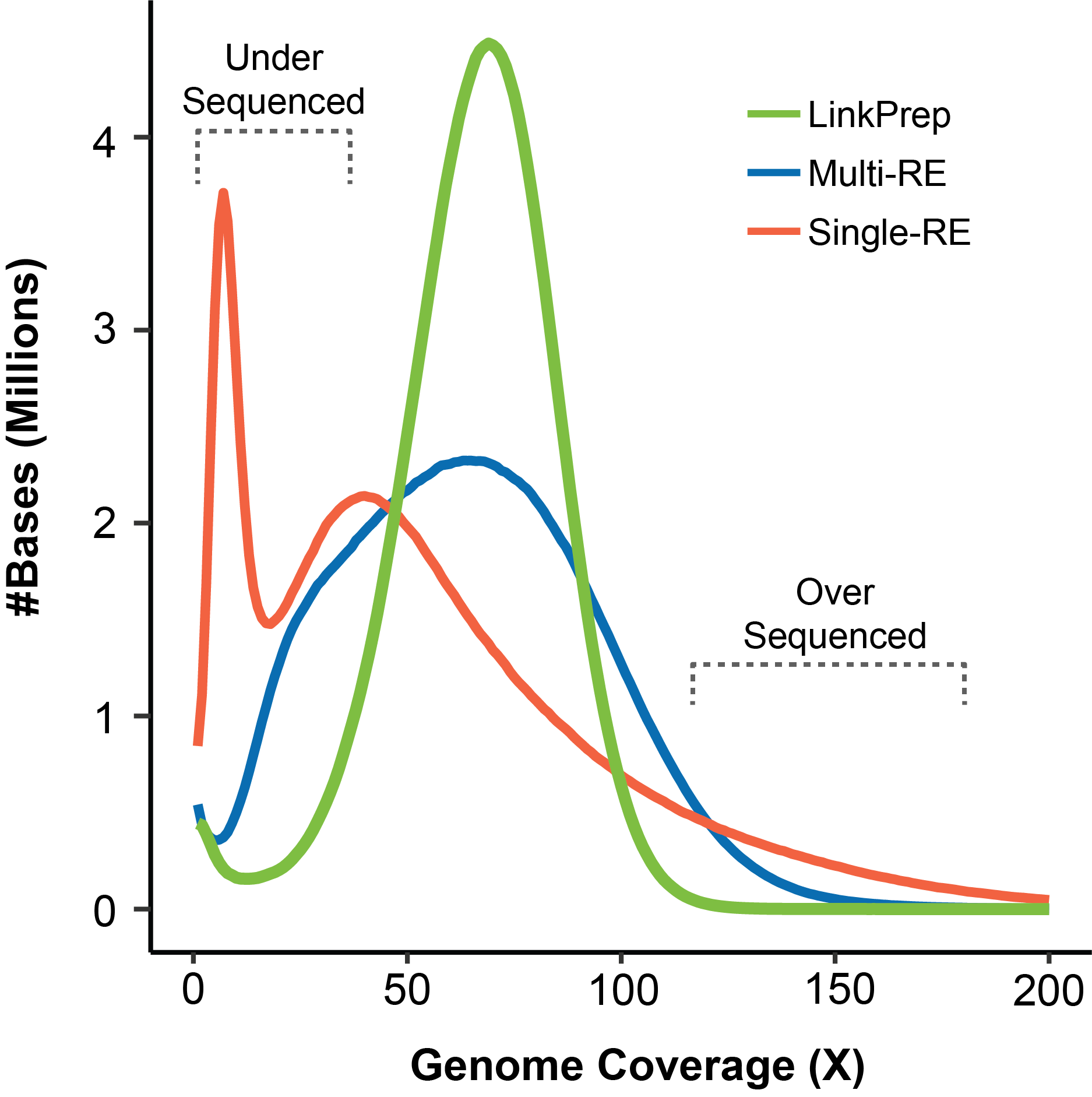
Uniform Coverage Across The Genome
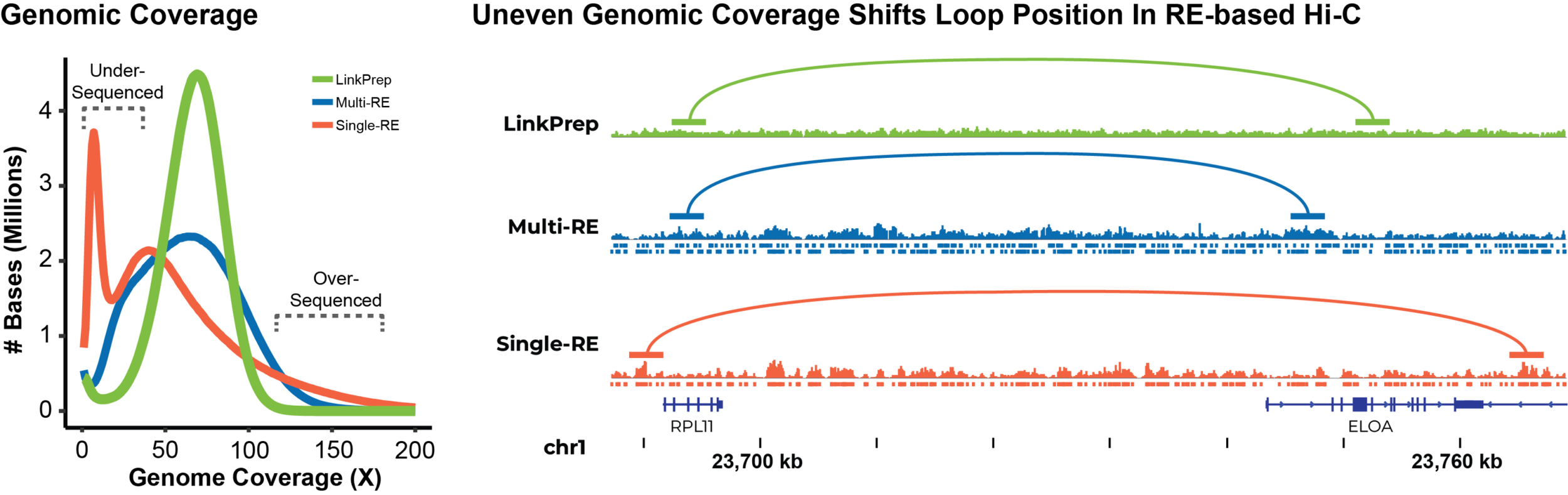
LinkPrep™ technology delivers uniform genome coverage compared to the wider non-Poisson coverage distribution of traditional Hi-C data due to restriction-enzyme based sequence bias.
The improved coverage (green curve) underscores the assay’s ability to achieve higher resolution and more accurate detection of structural variations and chromatin interactions. This capability offers a clearer understanding of complex genomic regions, enabling you to extract more meaningful insights from your data.
A Complete Solution - Sample to Insight
The Dovetail® Analysis Portal gives every Dovetail Genomics customer instant access to streamlined, cloud-based analysis of NGS FASTQ files from Dovetail® linked-read libraries. Choose from supported workflows for somatic variant calling (human) and epigenetic feature calling (human and mouse)—no bioinformatics setup required. Receive clear, ready-to-use results through an interactive summary report and a conveniently packaged downloadable archive.
The Dovetail® LinkPrep™ Kit includes all the critical components needed to take your projects from sample to sequencer with an easy and simplified workflow.
“I have found the Dovetail® LinkPrep™ Kit to be easy to use and not affected by tissue or cell morphology. It’s been very well suited to my study of looping interactions compared to the other kits I have used.”
Run LinkPrep™ in your lab
-
-
-
User Guides
-
Dovetail® LinkPrep™ Kit, 8 Samples, 8 Libraries
Includes:
LinkPrep™ Proximity Core 4°C
LinkPrep™ Proximity Core -20°C
LinkPrep™ Library Module for Illumina
LinkPrep™ Primer Module Set 1
Dovetail® Cell Isolation Module
Click to Explore Applications
Genetic Variation
3D Genomics
Genome Assembly
One Assay, Many Applications
Our innovative LinkPrep™ technology powers a wide range of applications including comprehensive detection of somatic genetic variation, 3D genome structure mapping, and the production of haplotype-aware genome assemblies.
Explore your application of interest through our application modules below
Genetic Variation
Improved whole genome de novo variant discovery with Dovetail® LinkPrep™ Technology
Unravel Oncogenic Drivers Through Comprehensive de novo Somatic Variation Detection
High-sensitivity detection of SVs, SNVs, InDels, and CNVs – completing your view of somatic variation.
How does LinkPrep support genetic variant detection?
Structural variants (SVs)—large insertions, deletions, translocations, and inversions—play a key role in tumor initiation, progression, and drug resistance
Yet they remain one of the most under-detected classes of somatic mutations due to the limitations of conventional sequencing:
Short-read WGS often fails to capture large SVs due to limited molecule length
False positives are common, requiring manual review and validation
High molecular weight DNA or specialized platforms are often required—incompatible with clinical workflows
As a result, many tumor samples are labeled “driver-negative”, even though structural drivers may be present but missed. That’s why we built Dovetail® LinkPrep assay paired with the Dovetail® Analysis Portal—a short-read-based, linked-read solution that empowers researchers to confidently detect structural variants alongside SNVs, InDels, and CNVs in a single assay.
Customer Testimonial
"Our lab is excited about the potential of LinkPrep™ data. We currently use a range of NGS-based assays to catalog genetic variations spanning from SNVs up to large SVs. LinkPrep™ has the potential to become a key tool in our oncology research, offering powerful capabilities for the de novo detection of the full spectrum of somatic variants that we focus on."
Brian Walker, Ph.D.
Professor of Medical and Molecular Genetics at Indiana University School of Medicine
Capture SVs Missed by Other Technologies—Without the Noise
User Data Example
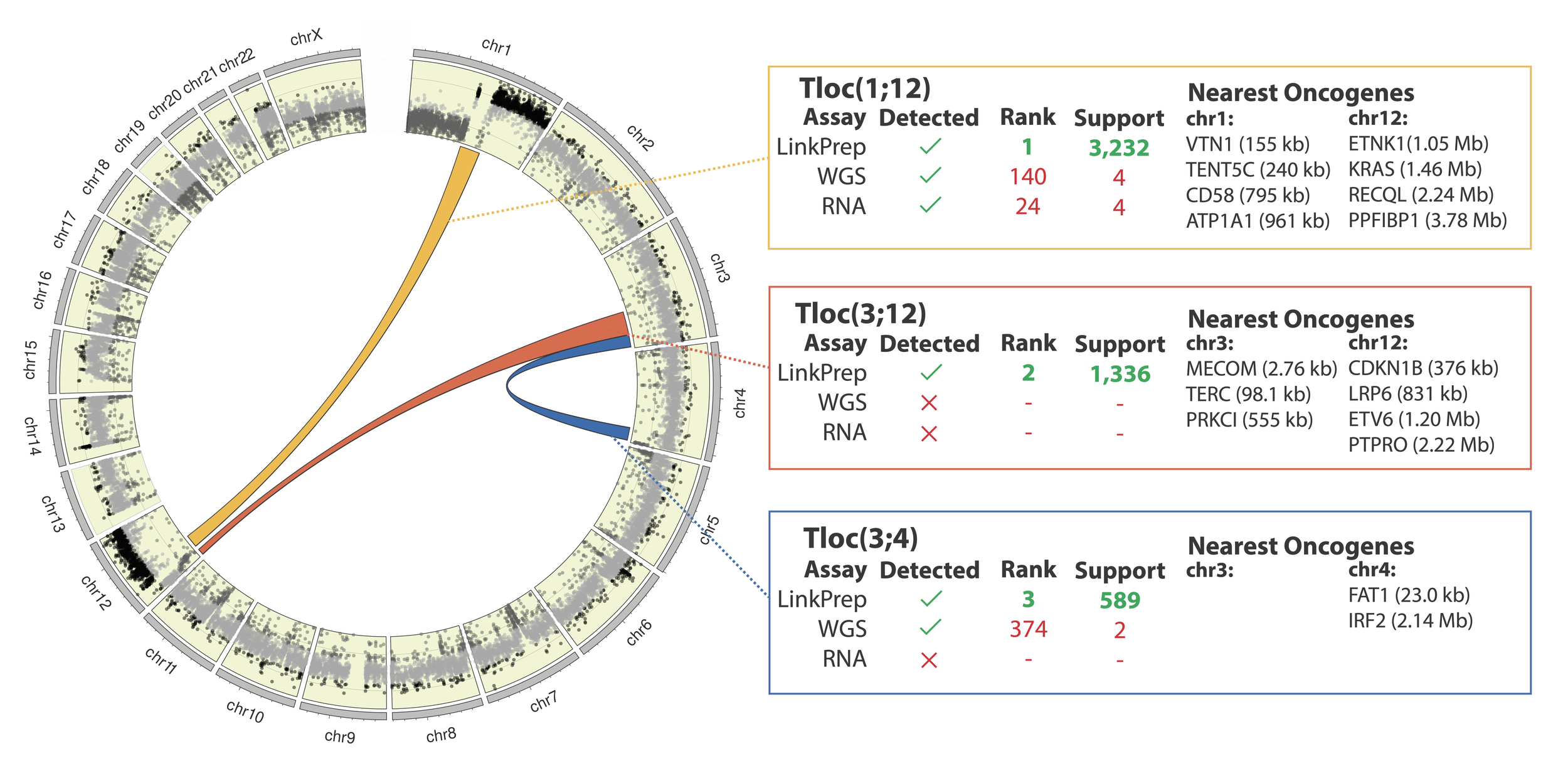
The LinkPrep assay, WGS and RNA-seq were used to characterize structural variants in a clinical ovarian sarcoma sample.
The LinkPrep™ assay accurately identified three distinct translocations without any false positives, supported by robust read counts (minimum read support >500).
These SVs involve several oncogenes known to be linked to ovarian cancer.
WGS and RNA-seq methods fall short, detecting only two and one translocations, respectively, with minimal read support (less than 4 reads).
These methods generated high numbers of false positives - 36 for RNA-seq and a staggering 402 for WGS - requiring extensive manual review and validation.
LinkPrep data excels at detecting genetic variants. Access highly sensitive SV detection with WGS-equivalent detection of SNVs, InDels, and CNVs, with LinkPrep technology, all on an Illumina platform. At its core the LinkPrep assay is an NGS solution that captures primary sequence information used to detect clinically relevant SNVs, ImDels, and CNVs. The unique chemistry behind the LinkPrep assay unlocks high sensitivity SV detection while minimizing false positives on your Illumina sequencer, even at low sequence depth.
K562 cells were profiled using both the LinkPrep Assay and WGS (sequenced to 30X coverage) and the results compared to a K562 truth set of known variants. The truth set was generated using a variety of technologies including WGS at >80X coverage, RNA-seq, long read sequencing. A) The LinkPrep data demonstrated WGS-like capabilities in detecting SNVs, InDels, and CNVs. B) The LinkPrep Assay demonstrated a much higher sensitivity for SVs compared to WGS. To further demonstrate the extreme sensitivity of SVs, LinkPrep data was down sampled to 0.01X genomic coverage.
Explore our oncology customer highlight
3D Genomics
Revolutionize your understanding of gene regulation and biological processes through the power of 3-dimensional DNA organization.
What does Dovetail® LinkPrep™ provide for 3D genomics?
From disease drivers to therapy:
Applying 3D genomics–powered insights helps our clients better understand, monitor, and characterize disease. Identify novel drivers across diverse phenotypes, including cancer, neurological disorders, autoimmune diseases, and more.
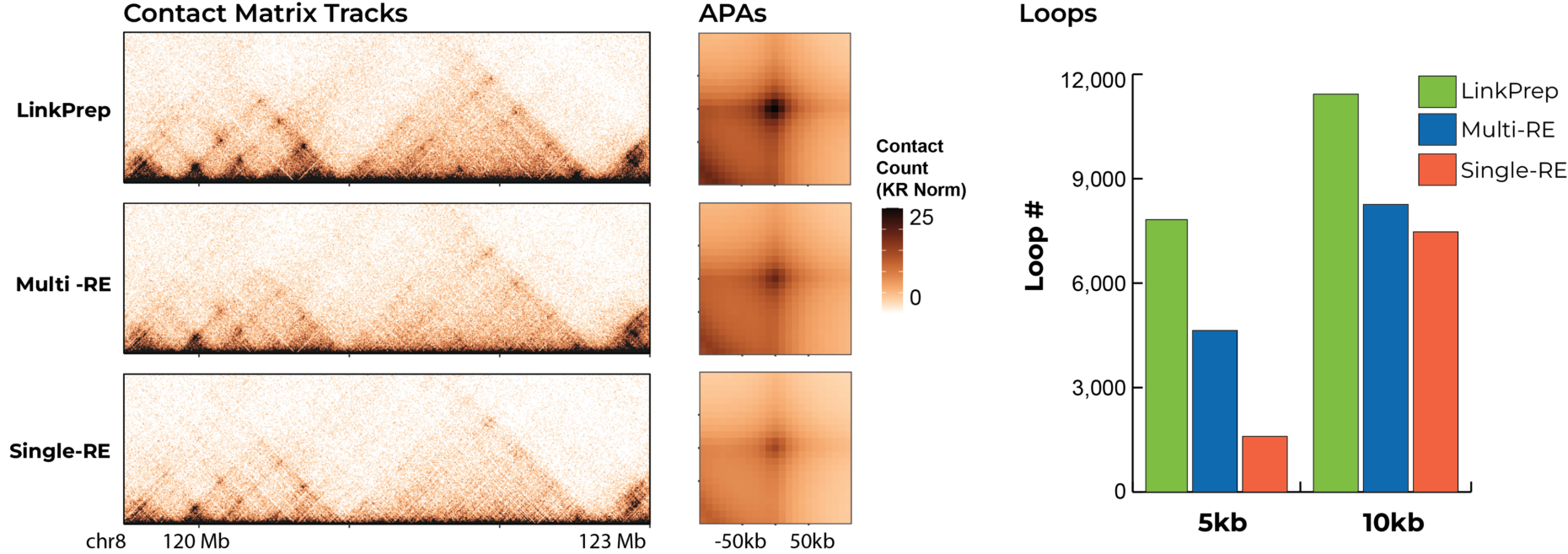
LinkPrep’s high data quality enables detection of more features, such as TADs and loops, than traditional Hi-C, even at lower sequencing depths. This efficiency reduces the number of libraries and total sequencing required, simplifying experimental design and execution.
Dovetail® LinkPrep™ provides unbiased, uniform genomic coverage for richer contact maps and more chromatin loop detection than traditional Hi-C. Its consistent sequence coverage ensures the most accurate chromatin interaction map.
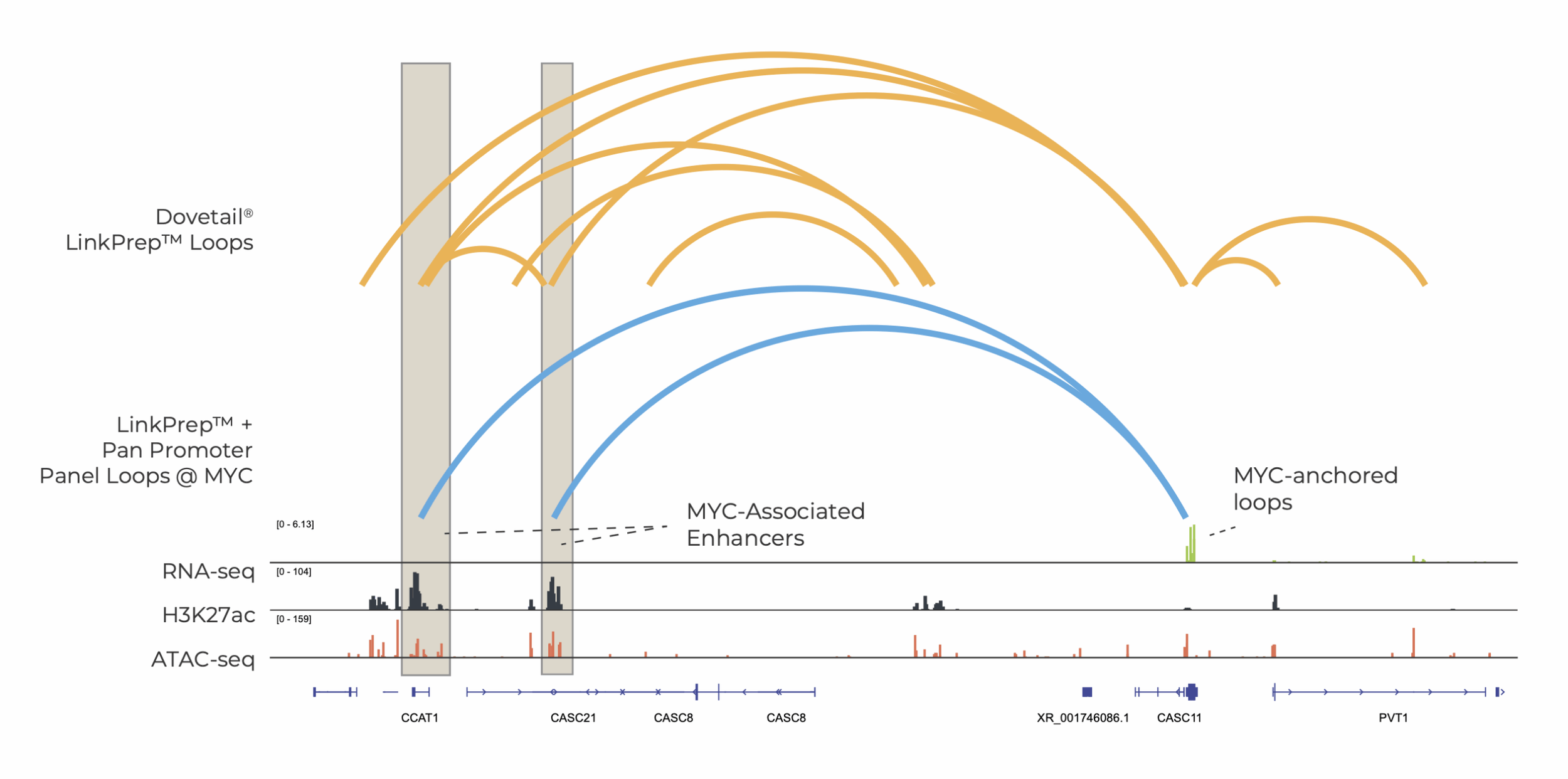
Not after genome-wide data? Focus on your target region.
LinkPrep is now compatible with hybrid capture
The Dovetail® LinkPrep™ Assay now supports hybrid capture. When paired with the Dovetail® Pan Promoter Panel, targeted LinkPrep™ data enables efficient promoter-anchored interaction profiling and enriches biologically meaningful signals, revealing connections between promoters, regulatory elements, and non-coding variants.
This powerful approach can accelerate research in GWAS, oncology, and 3D genomics. Whether you're using our Pan Promoter Panel, an off-the-shelf third-party panel, or your own custom design, targeting LinkPrep interactions delivers high-quality data, streamlined analysis, and scalability—all while maintaining the speed, accuracy, and affordability of the core assay.
Shop Full Solutions
Pan Promoter for LinkPrep™
Produce highly uniform sequence coverage across all parts of the genome.
Where the non-uniform restriction enzyme site distribution associated with traditional Hi-C methods often skews signals, you can be confident in the position of interactions detected by LinkPrep.
Customer Testimonial
“I have found the Dovetail® LinkPrep™ Kit to be easy to use and not affected by tissue or cell morphology. It’s been very well suited to my study of looping interactions compared to the other kits I have used.”
Benjamin Lebeau
Research Assistant, Nanyang Technological University
Genome Assembly
Reduce Time and Improve Genome Assembly Quality with the Dovetail® LinkPrep™ Assay
How does LinkPrep™ enable genome assembly?
The Dovetail LinkPrep™ Assay streamlines the genome assembly process by rapidly generating Hi-C-like libraries essential for genome scaffolding. All users with standard molecular biology training can access the assay due to the simplicity of the workflow. The kit is configured to support all steps from sample to sequencing library.
Rapid workflow – sample to library in one day.
Simple to use – accessible to novice users.
Low sample input – requires as little as 5 mg of tissue.
No sonication required - specialized equipment not necessary.
Automation compatible – amenable to high-throughput settings.
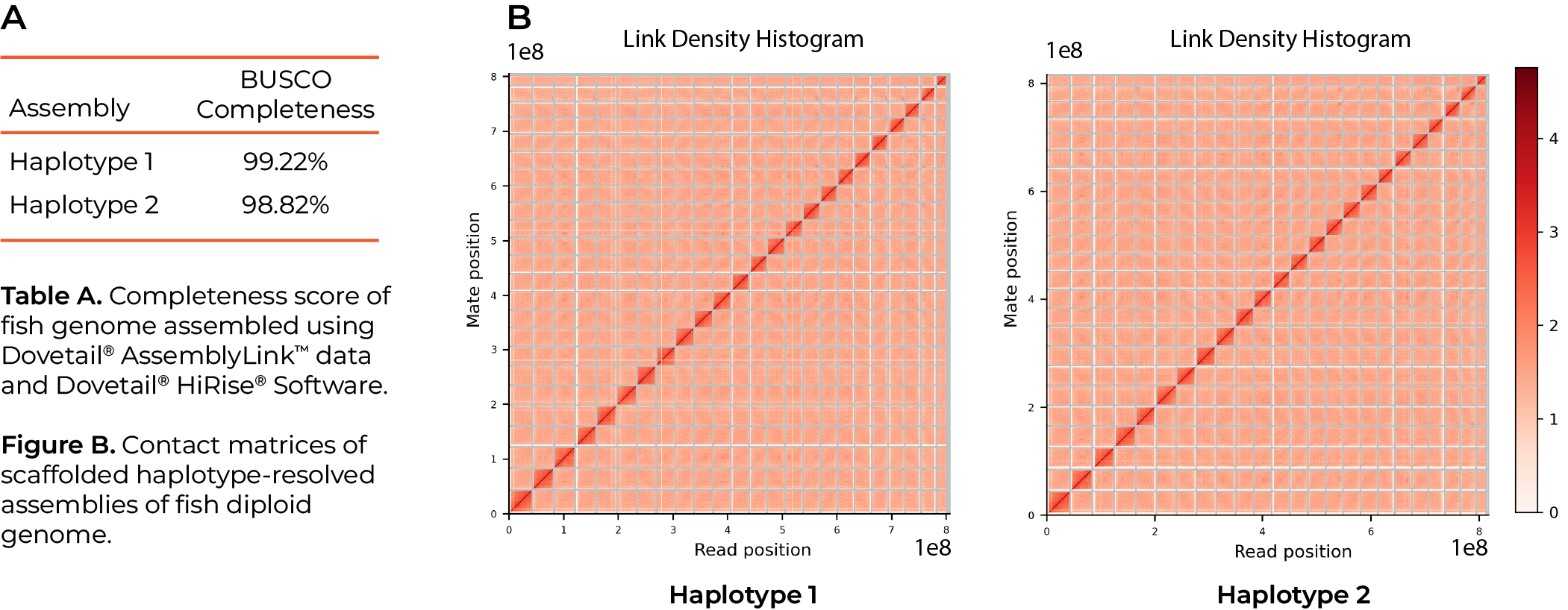
Produce Haplotype Resolved Assemblies
The LinkPrep kit offers a faster workflow that does not compromise on data quality. The assay delivers highly uniform sequence coverage facilitating variant phasing critical to producing true haplotype resolved assemblies. Moreover, this unique attribute leads to an easier path to chromosome-scale assembly of higher ploidy genomes.
High quality data – supports chromosome scale assembly.
Capture phase information – enables haplotype resolved and polyploid genome assembly.
Uniform sequence coverage – lowers sequencing depth requirements.
Customer Testimonial
"We're thrilled to onboard the Dovetail LinkPrep™ kit. The Minderoo OceanOmics Centre at UWA is using Dovetail Hi-C preps to scaffold whole genome assemblies of marine vertebrate species, as we create a comprehensive library of high-quality reference genomes. The refined workflow will decrease hands-on time in the lab and increase sequencing throughput. The reduced input requirements from fresh frozen animal tissue means we can now tackle even the smallest specimens without compromising sequencing quality or complexity."
Adrianne Doran,
Scientific Officer, Minderoo OceanOmics Centre at UWA
Customer Testimonial
"We are very excited by the capabilities of the AssemblyLink Kit. It takes just one day to go from sample preparation to sequencing for a novel de novo assembly or for high-resolution 3D genomic interactions. This efficiency will have a great impact, cutting down the process from three days to one without compromising on deliverables. At the Australian Genome Research Facility, where we strive to provide premium customer services, we can't wait to test it further."
Dr. Dhanya Sooraj, Senior Scientist
Innovation and Development, Australian Genome Research Facility