Menu
Menu

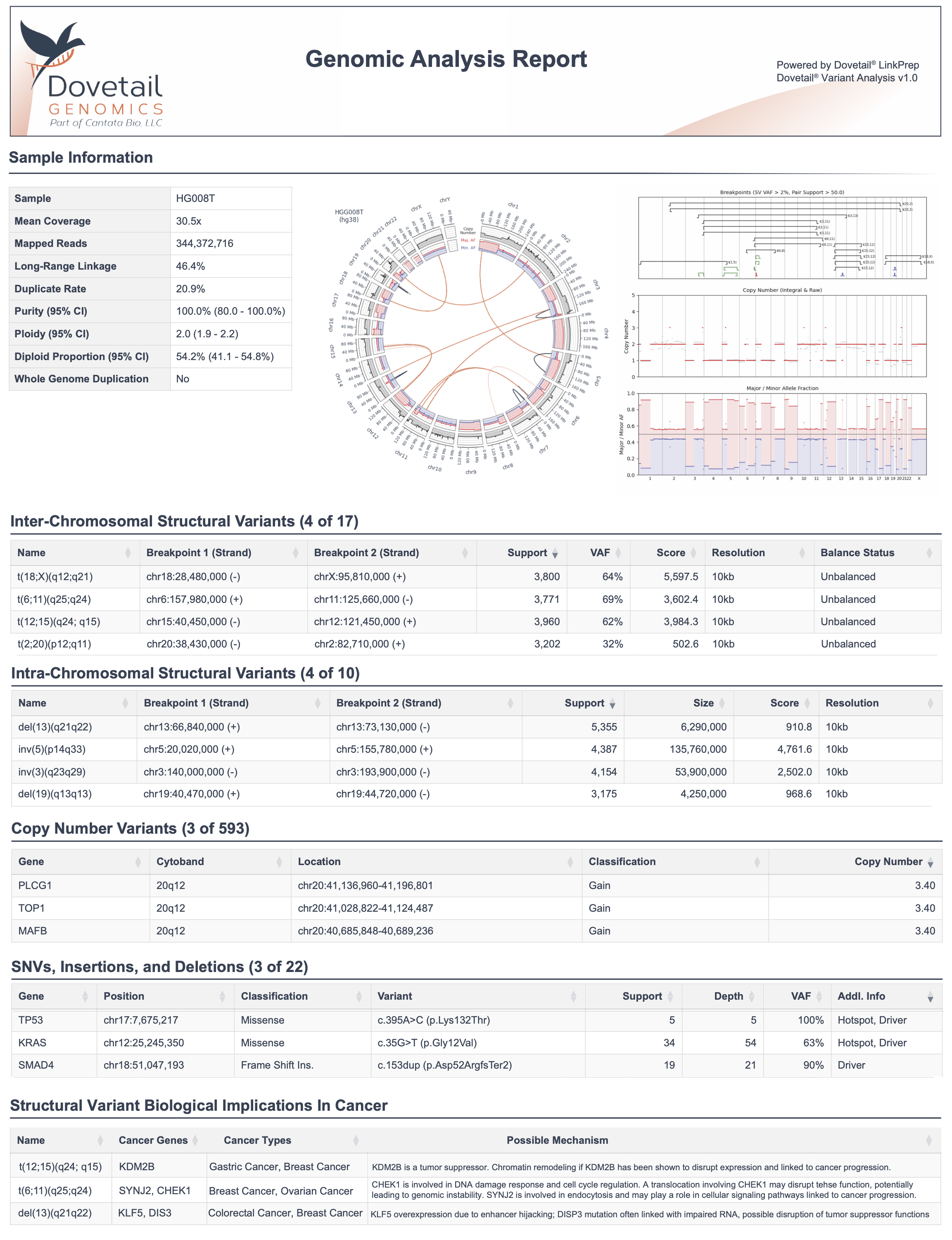
Structural variants (SVs)—large insertions, deletions, translocations, and inversions—play a key role in tumor initiation, progression, and drug resistance
Yet they remain one of the most under-detected classes of somatic mutations due to the limitations of conventional sequencing:
As a result, many tumor samples are labeled “driver-negative”, even though structural drivers may be present but missed. That’s why we built Dovetail® LinkPrep assay paired with the Dovetail® Analysis Portal—a short-read-based, linked-read solution that empowers researchers to confidently detect structural variants alongside SNVs, InDels, and CNVs in a single assay.


Dovetail® LinkPrep Technology extends the capabilities of short-read sequencers by capturing long-range genomic information. When paired with the Dovetail® Analysis Portal, you gain a simplified, end-to-end workflow for somatic variant discovery—without specialized informatics or equipment.
"Our lab is excited about the potential of LinkPrep™ data. We currently use a range of NGS-based assays to catalog genetic variations spanning from SNVs up to large SVs. LinkPrep™ has the potential to become a key tool in our oncology research, offering powerful capabilities for the de novo detection of the full spectrum of somatic variants that we focus on."
Brian Walker, Ph.D.
Professor of Medical and Molecular Genetics, University of Miami Miller School of Medicine
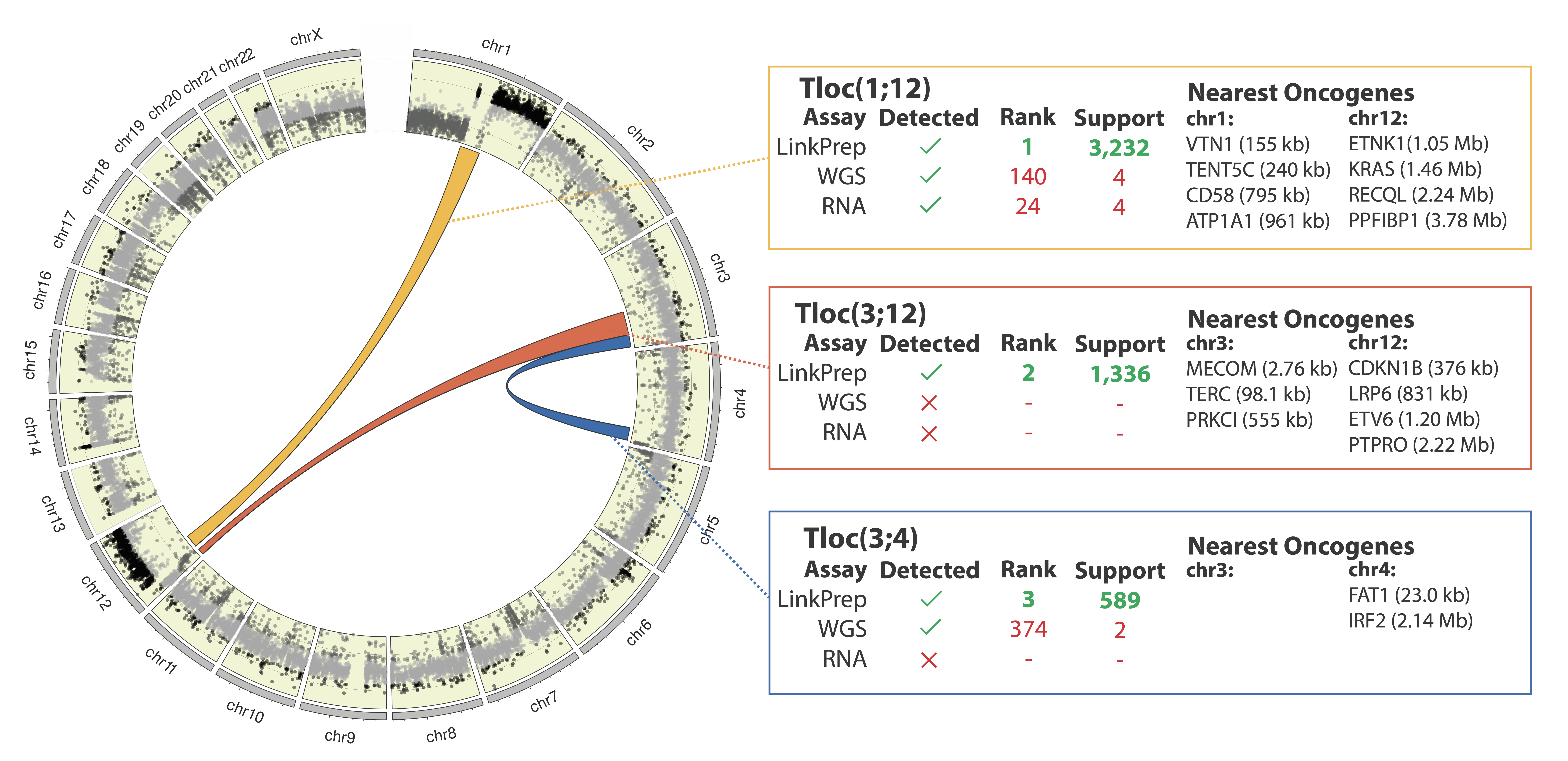
The LinkPrep assay, WGS and RNA-seq were used to characterize structural variants in a clinical ovarian sarcoma sample.
 100 Enterprise Way
100 Enterprise Way