Menu
Menu

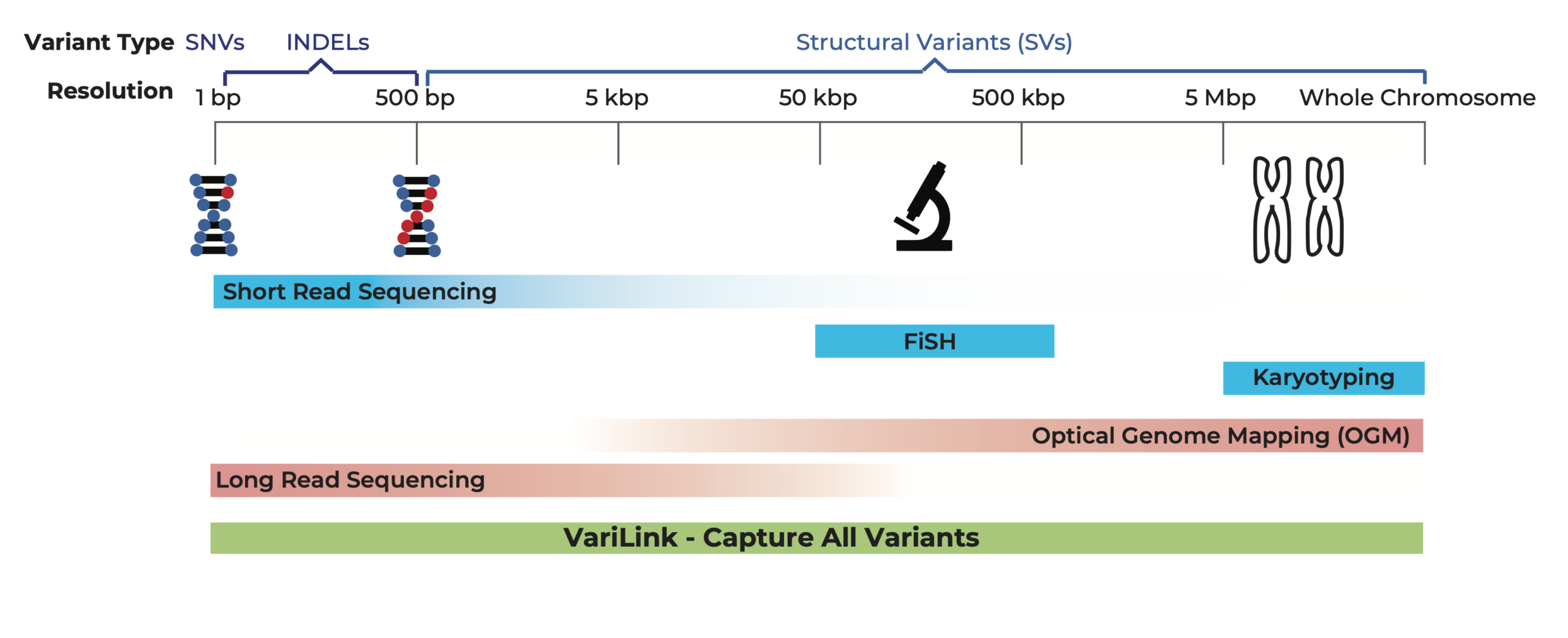
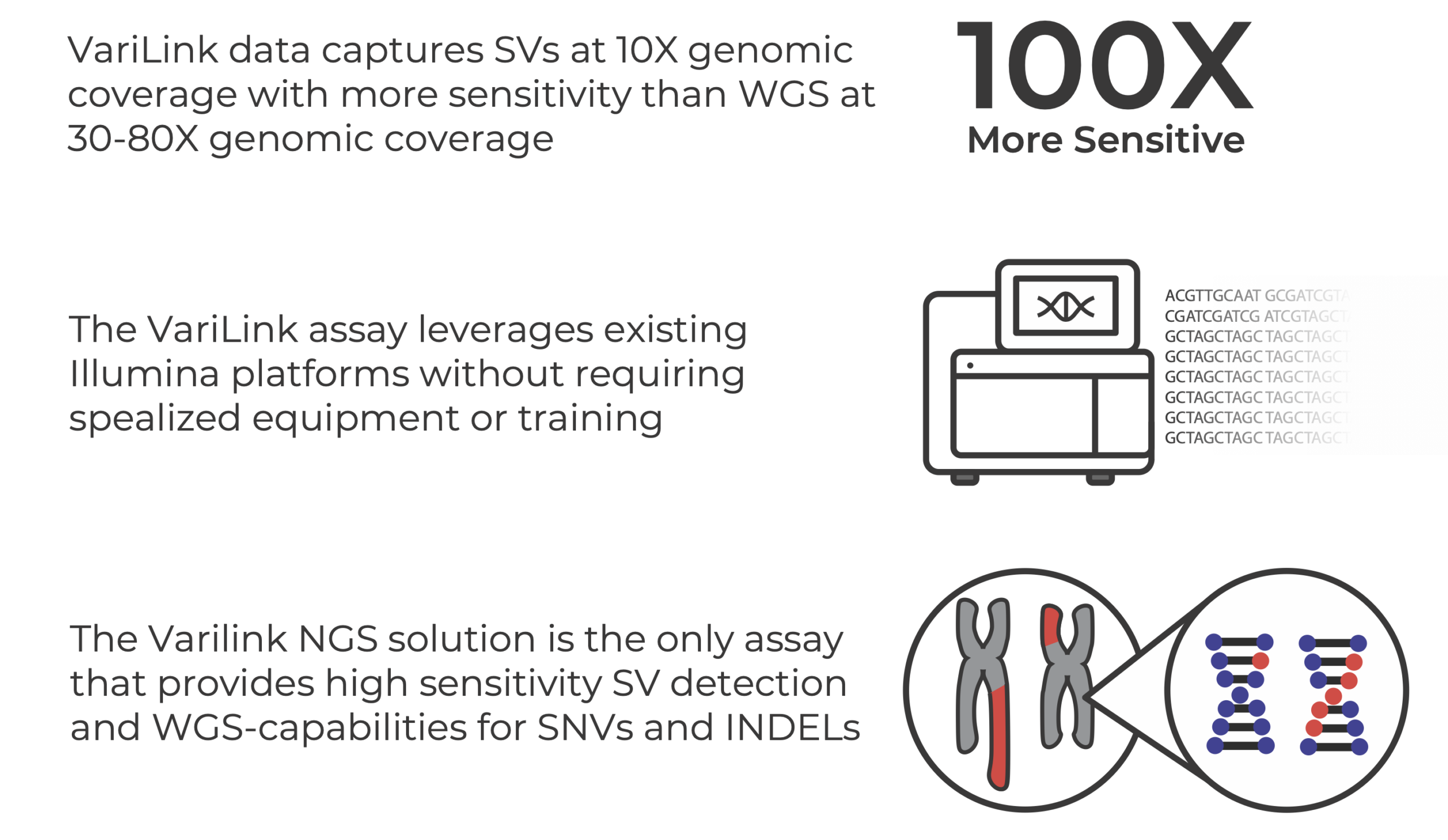
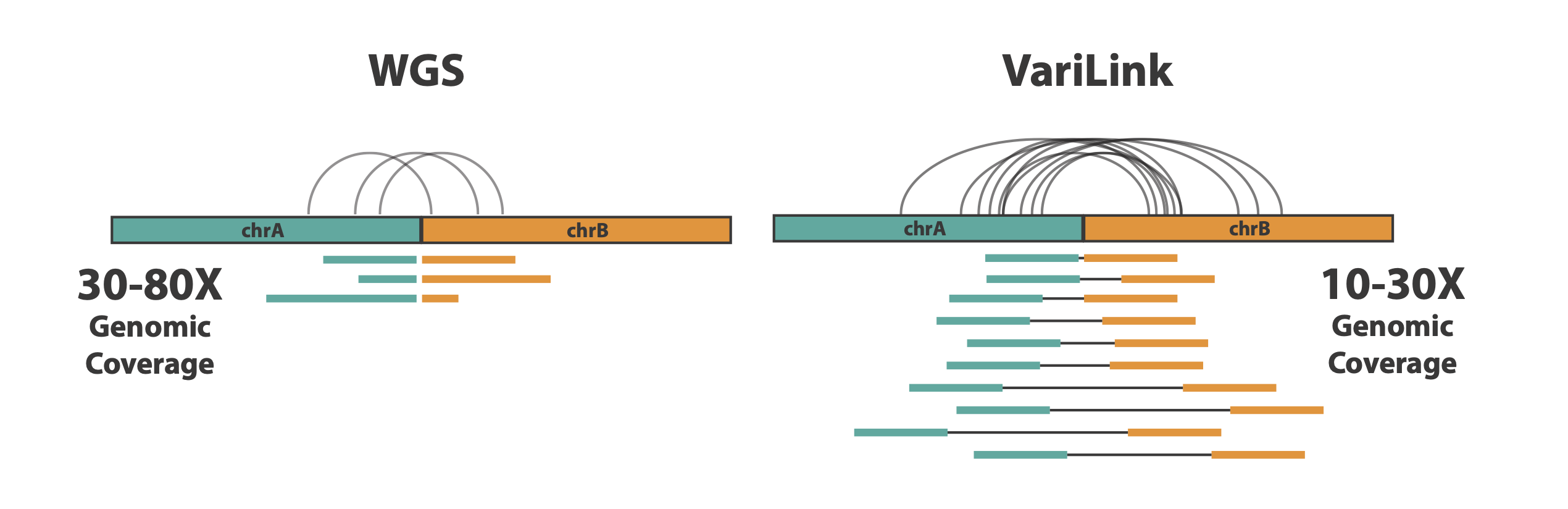
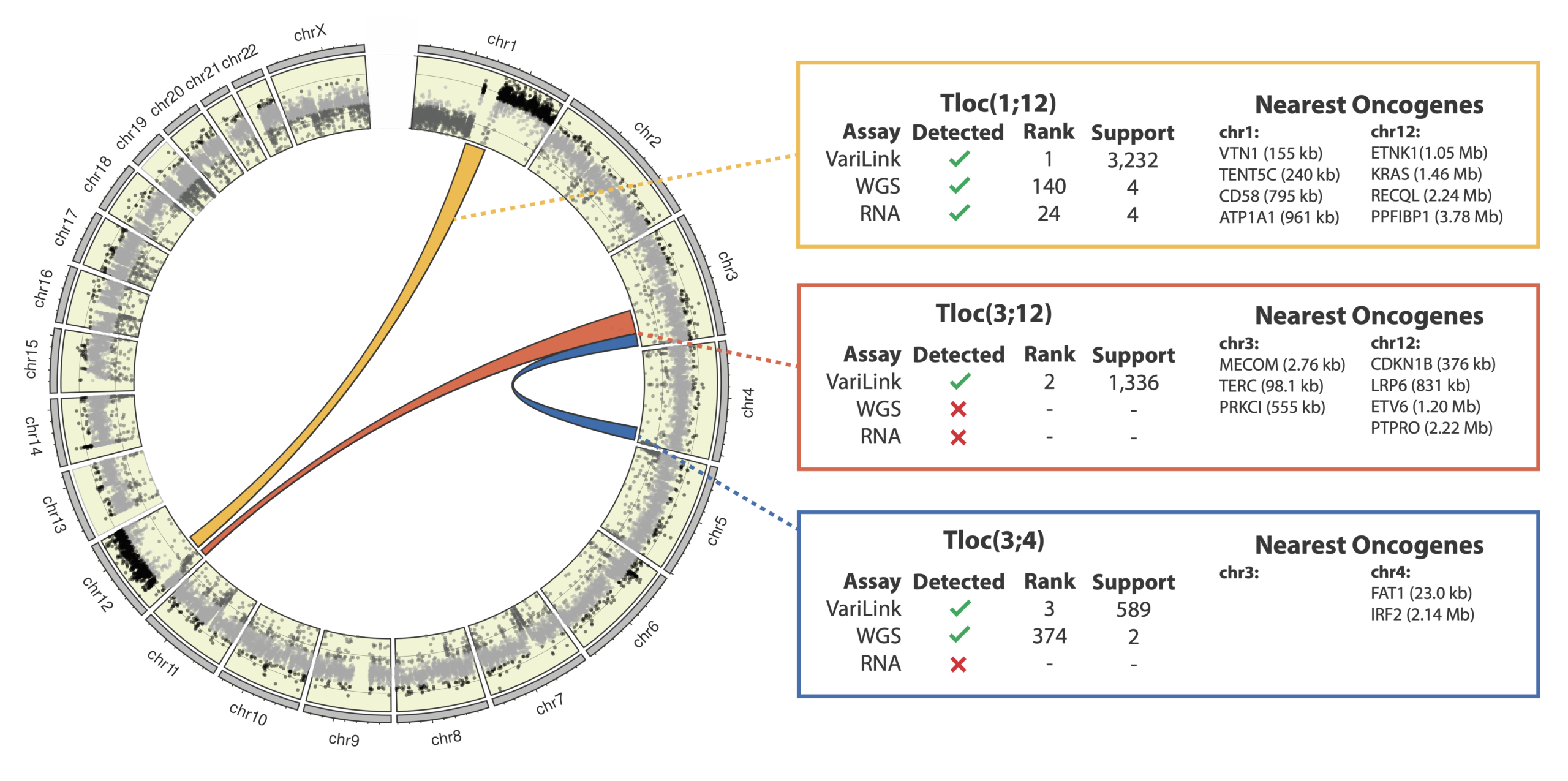
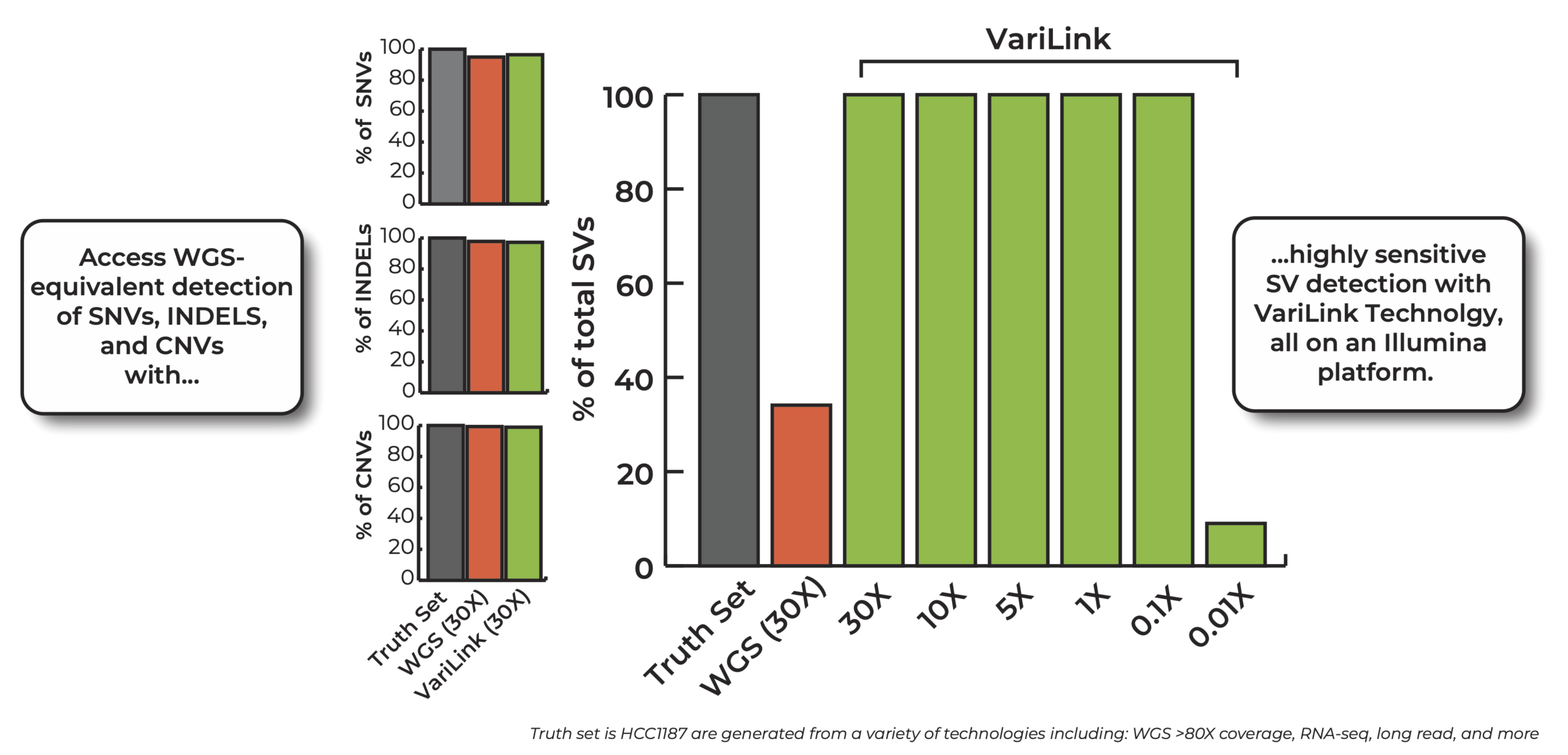
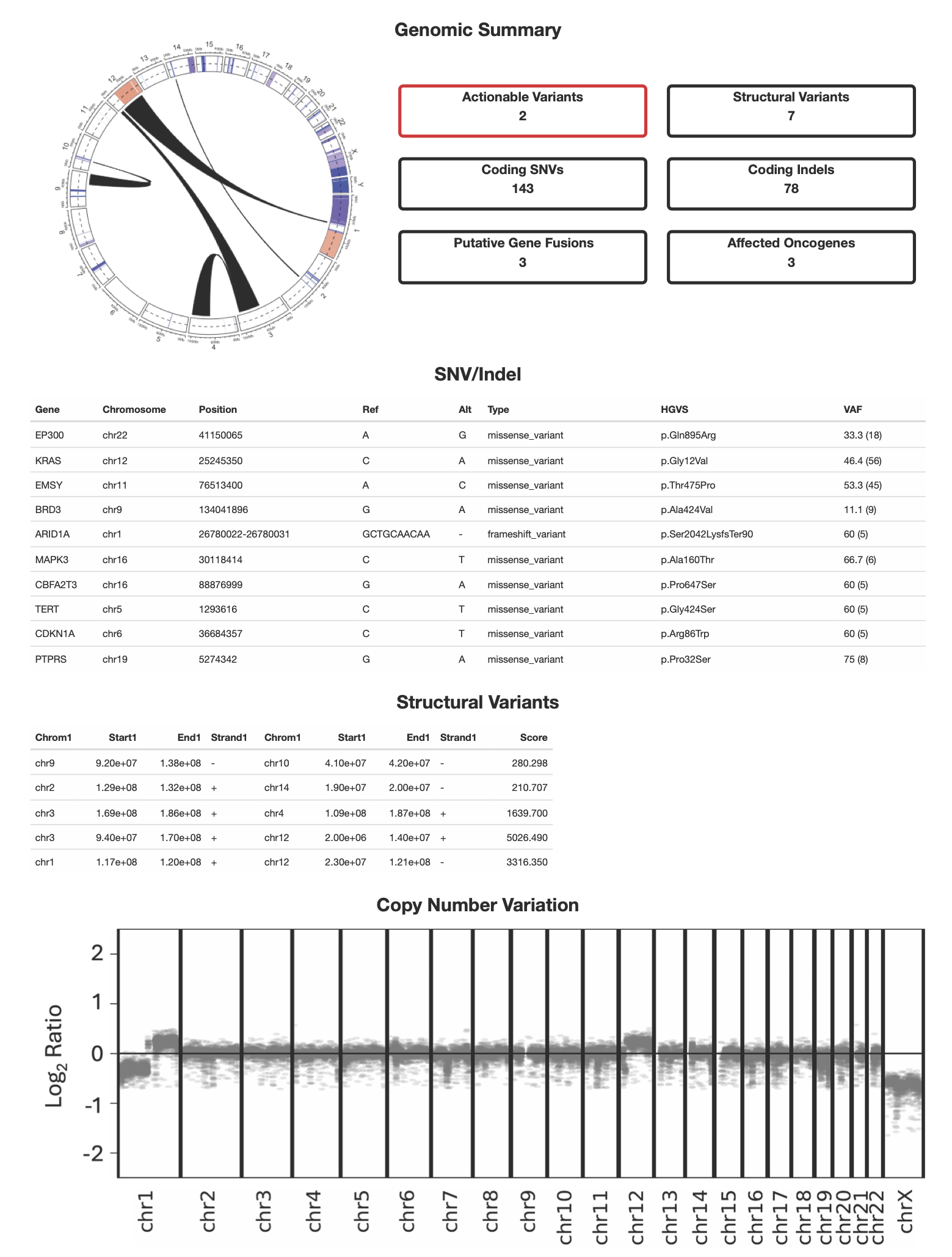
Current technologies have significant gaps in detecting the range of genetic variation that drive cancers. Structural variation, in particular, is incredibly diverse and complex, and is a challenge to detect with conventional WGS methods. The Dovetail® VariLink™ Assay allows for the highly sensitive and accurate detection of structural variation, while preserving the capabilities of short read sequencing in detection of SNVs, Indels and CNV.
 100 Enterprise Way
100 Enterprise Way